abYsis
Library Module
Design antibody Library using Frequencies extracted from abYsis
abYsis Library Module
This latest addition allows researchers to design libraries where the residues to be included at selected positions are based on position specific frequencies observed in the database.
Choose which positions you want to vary, selecting either the single most frequent amino-acid for that position or a range of amino-acids whose inclusion is determined by adjusting the slider bars (choose between region or residue-position specific).
When complete export to excel for incorporating into a library order with your favoured provider.
Select Regions or Residues
Simply click on a whole CDR region or an individual residue to toggle selection. Then choose what frequency selection your would like.
Select Organism
Use the Organism menu to restrict frequencies to a single organism of interest.
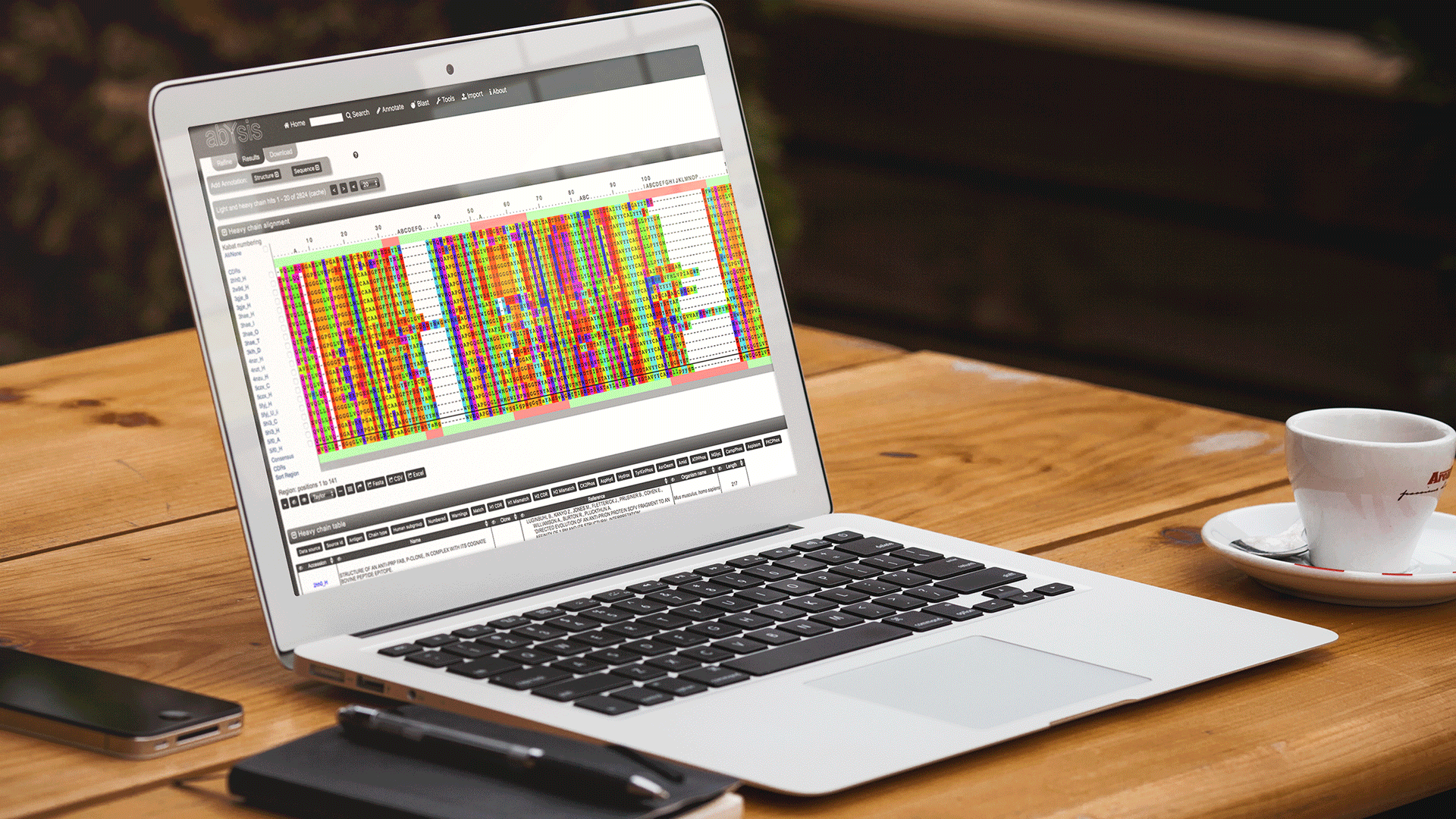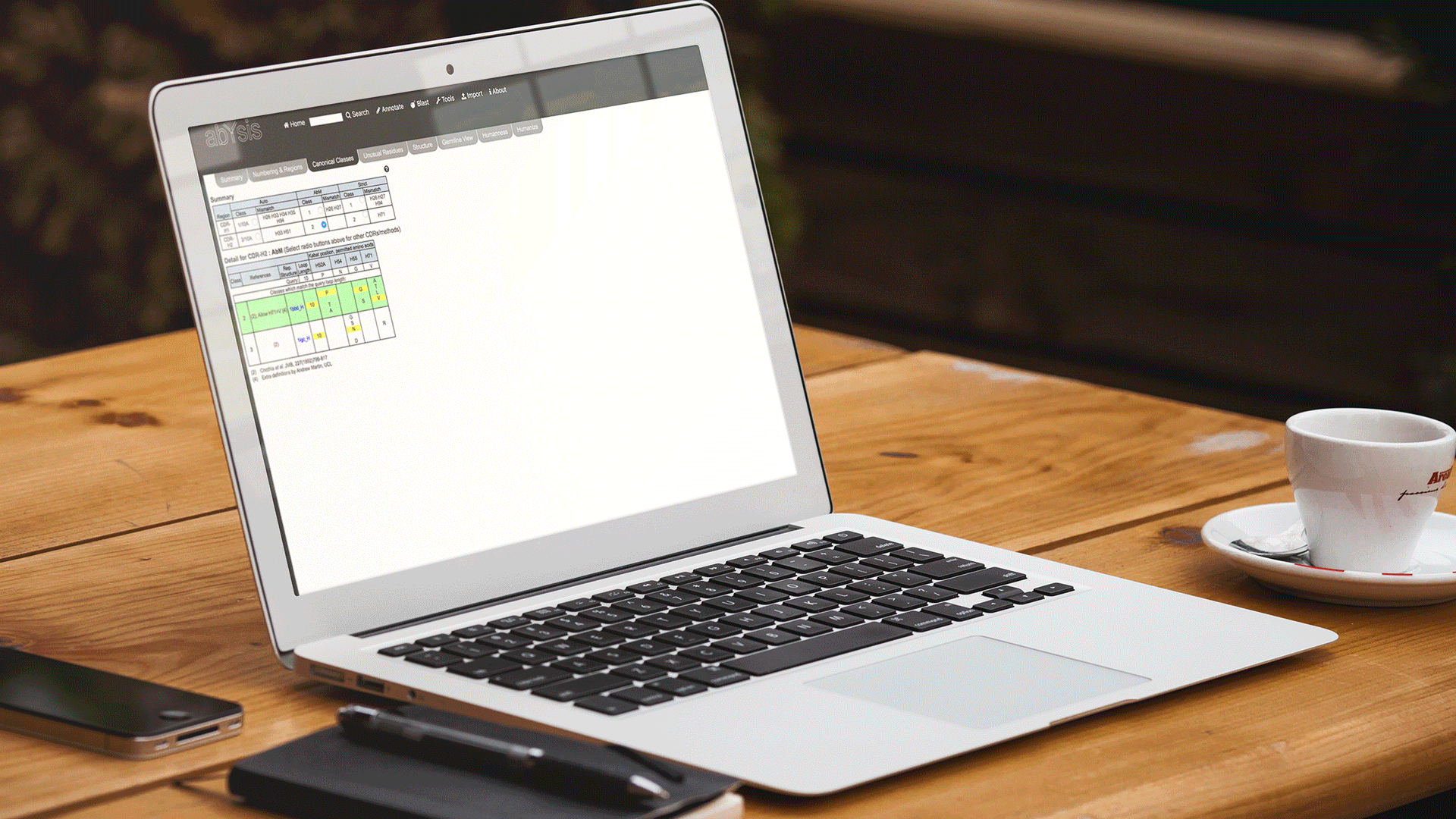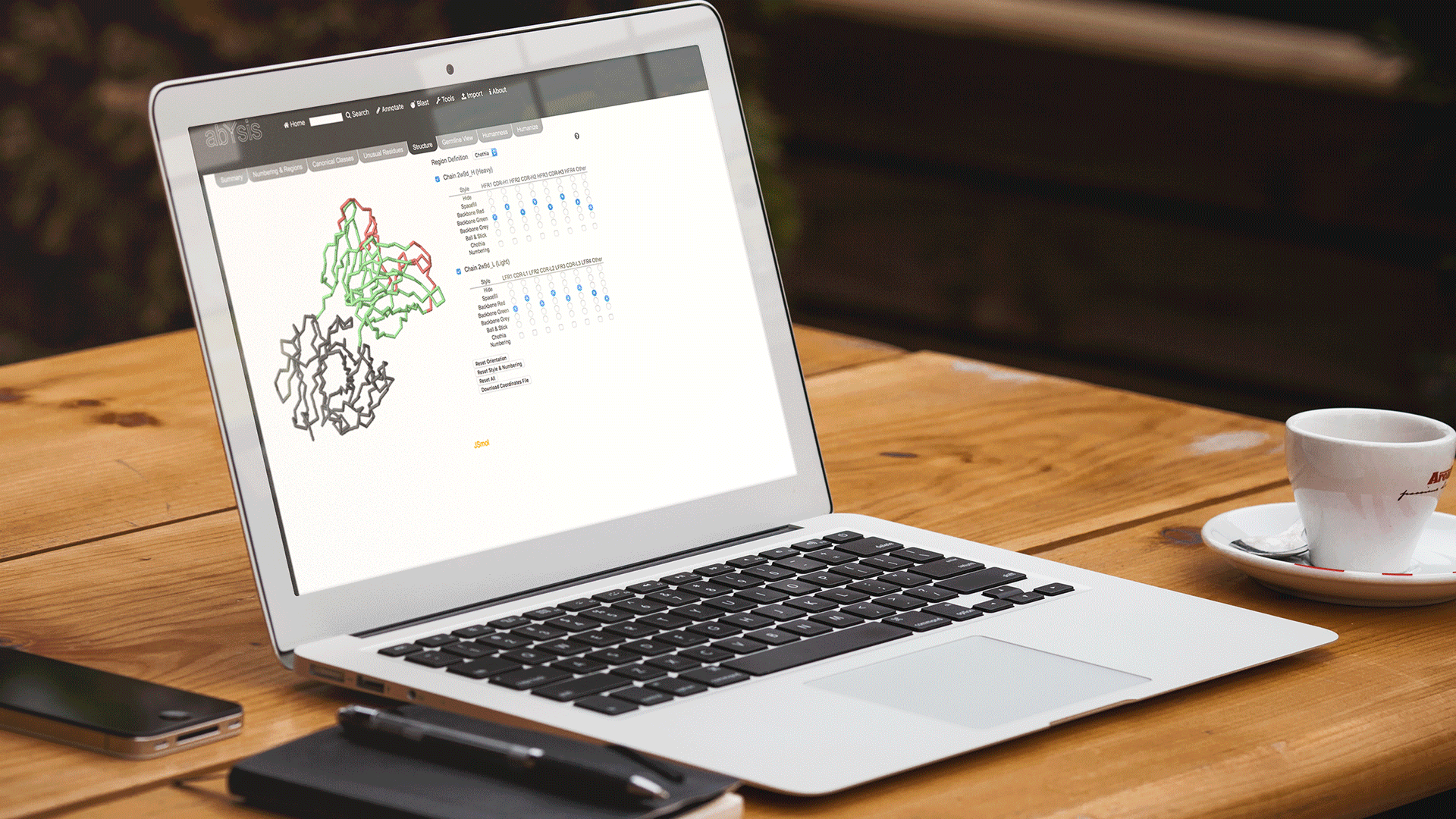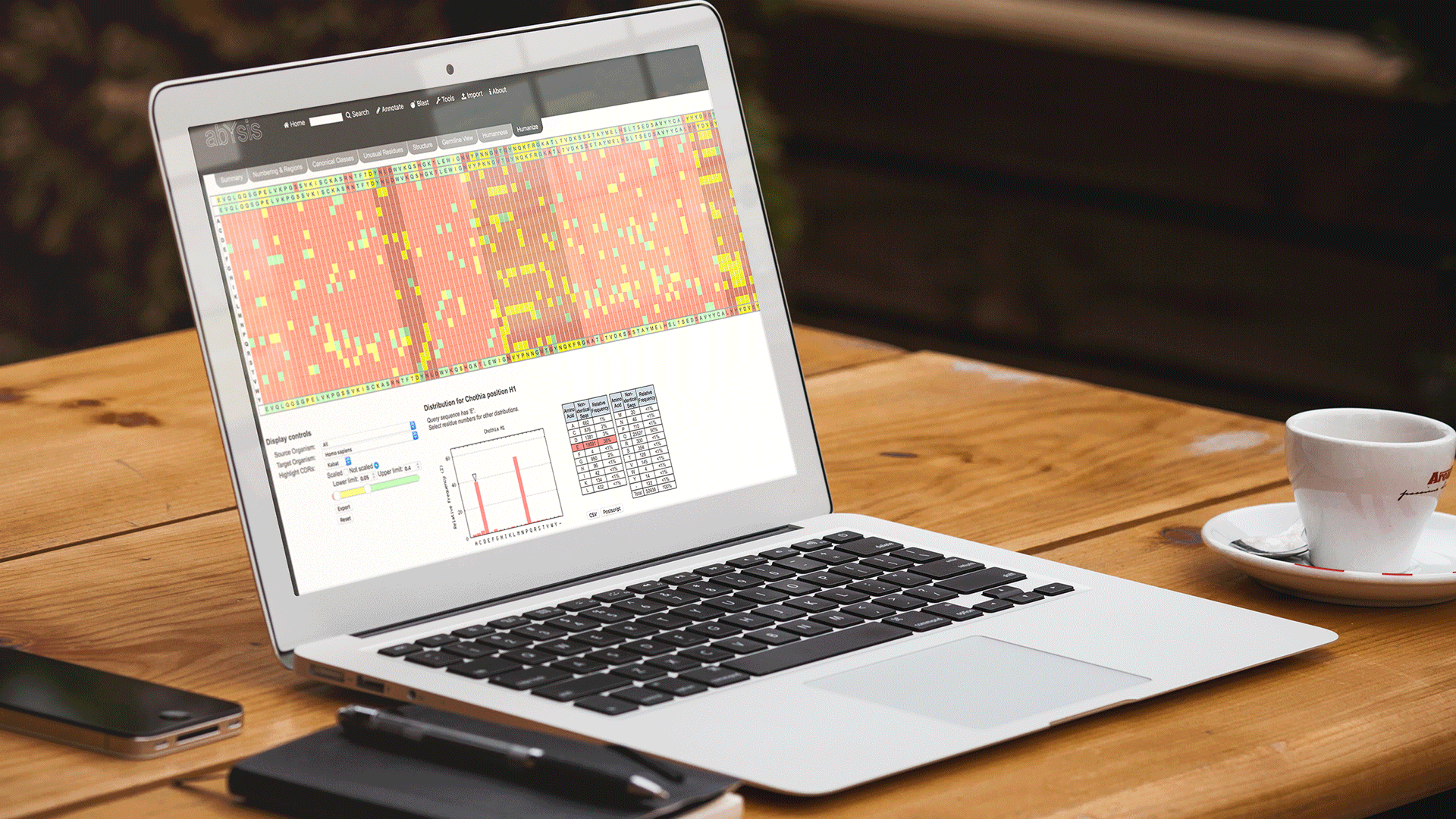
Frequencies
Either select the most frequently occuring residue at a position (tick) or deselect the tick box and move the slider to dynamically adjust the number of amino-acids that fall above your user-defined threshold.
Export
Export the results to Excel once finished. Each residue will have its % frequency shown.
